DNA tests of Lassa virus mid-outbreak helped Nigeria target its response
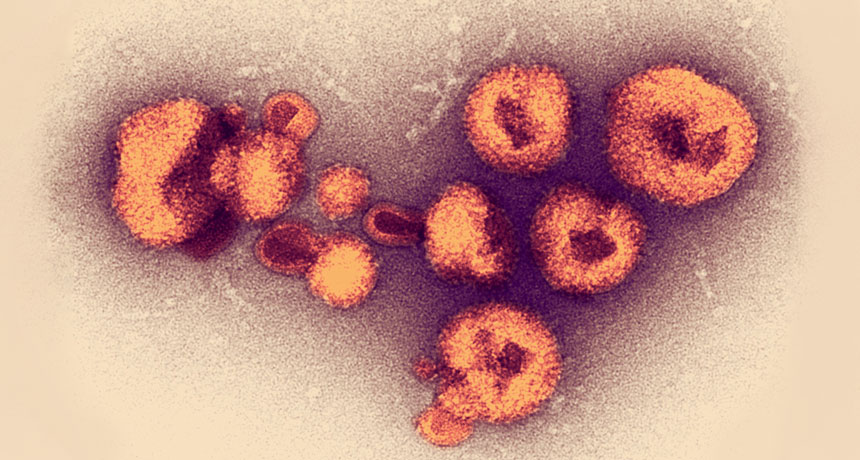
When an outbreak of a viral hemorrhagic fever hit Nigeria in 2018, scientists were ready: They were already in the country testing new disease-tracking technology, and within weeks managed to steer health workers toward the most appropriate response.
Lassa fever, which is transmitted from rodents to humans, pops up every year in West Africa. But 2018 was the worst season on record for Nigeria. By mid-March, there were 376 confirmed cases — more than three times as many as by that point in 2017 — and another 1,495 suspected. Health officials weren’t sure if the bad year was being caused by the strains that usually circulate, or by a new strain that might be more transmissible between humans and warrant a stronger response.
New technology for analyzing DNA in the field helped answer that question mid-outbreak, confirming the outbreak was being caused by pretty much the same strains transmitted from rodents to humans in past years. That rapid finding helped Nigeria shape its response, allowing health officials to focus efforts on rodent control and safe food storage, rather than sinking time and money into measures aimed at stopping unlikely human-to-human transmission, researchers report in the Jan. 4 Science.
While the scientists were reporting their results to the Nigeria Centre for Disease Control, they were also discussing the data with other virologists and epidemiologists in online forums. This kind of real-time collaboration can help scientists and public health workers “see the bigger picture about pathogen spread,” says Nicholas Loman, a microbial genomicist at the University of Birmingham in England who was not involved in the research.
Portable DNA sequencers, some as small as a cell phone, have allowed scientists to read the genetic information of viruses emerging in places without extensive lab infrastructure. Looking for genetic differences between patient samples can give clues to how a virus is being transmitted and how quickly it’s changing over time — key information for getting outbreaks under control. If viral DNA from several patients is very similar, that suggests the virus may be transmitted between people; if the DNA is more distinct, people might be picking up the virus independently from other animals.
The technology has also been used amid recent Ebola and Zika outbreaks. But the Lassa virus presents a unique challenge, says study coauthor Stephan Günther, a virologist at the Bernhard-Nocht-Institute for Tropical Medicine in Hamburg, Germany. Unlike Ebola or Zika, Lassa has a lot of genetic variation between strains. So while the same small regions of DNA from various strains of Ebola or Zika can be identified for analysis, it’s hard to accurately target similar regions for comparison among Lassa strains.
Instead, Günther and his team used a tactic called metagenomics: They collected breast milk, plasma and cerebrospinal fluid from patients and sequenced all the DNA within — human, viral and anything else lurking. Then, the team picked out the Lassa virus DNA from that dataset.
All told, the scientists analyzed Lassa virus DNA from 120 patients, far more than initially intended. “We went to the field to do a pilot study,” Günther says. “Then the outbreak came. And we quickly scaled up.” Preexisting relationships in Nigeria helped make that happen: The team had been collaborating for a decade with researchers at the Irrua Specialist Teaching Hospital and working alongside the World Health Organization and the Nigeria Centre for Disease Control.
Analyzing and interpreting the massive amounts of data generated by the metagenomics approach was a challenge, especially with limited internet connection, Günther says. Researchers analyzed 36 samples during the outbreak — less than a third of their total dataset, but still enough to guide the response. The full analysis, completed after the outbreak, confirmed the initial findings.
A metagenomics approach could be useful in disease surveillance more broadly. Currently, “we look for things that we know about and expect to find. Yet evidence from Ebola in West Africa and Zika in the Americas is that emerging pathogens can pop up in unexpected places, and take too long to be recognized,” Loman says. Sequencing all DNA in a sample, he says, could allow scientists to detect problem pathogens before they cause outbreaks.Instead, Günther and his team used a tactic called metagenomics: They collected breast milk, plasma and cerebrospinal fluid from patients and sequenced all the DNA within — human, viral and anything else lurking. Then, the team picked out the Lassa virus DNA from that dataset.
All told, the scientists analyzed Lassa virus DNA from 120 patients, far more than initially intended. “We went to the field to do a pilot study,” Günther says. “Then the outbreak came. And we quickly scaled up.” Preexisting relationships in Nigeria helped make that happen: The team had been collaborating for a decade with researchers at the Irrua Specialist Teaching Hospital and working alongside the World Health Organization and the Nigeria Centre for Disease Control.
Analyzing and interpreting the massive amounts of data generated by the metagenomics approach was a challenge, especially with limited internet connection, Günther says. Researchers analyzed 36 samples during the outbreak — less than a third of their total dataset, but still enough to guide the response. The full analysis, completed after the outbreak, confirmed the initial findings.
A metagenomics approach could be useful in disease surveillance more broadly. Currently, “we look for things that we know about and expect to find. Yet evidence from Ebola in West Africa and Zika in the Americas is that emerging pathogens can pop up in unexpected places, and take too long to be recognized,” Loman says. Sequencing all DNA in a sample, he says, could allow scientists to detect problem pathogens before they cause outbreaks.